Highlights from our recently published manuscript on Mycobacterium pinnipedii1
By A.M. Guimaraes and T.T. Silva-Pereira
Why did we perform this study?
 Initiating a study exclusively on M. pinnipedii came as a surprise for us. As part of our Morris Animal Foundation Grant, our aim was to explore the genetic diversity of the Mycobacterium tuberculosis complex as a whole. To achieve this objective, we needed to sequence the genome of M. pinnipedii, because it had never been sequenced before. Our collaborators from the Laboratory of Bacterial Zoonosis at the College of Veterinary Medicine from the University of São Paulo had previously isolated two strains of M. pinnipeddi (MP1 and MP2) from a stranded sea lion carcass found in Capão da Canoa, Rio Grande do Sul (see map above)2. These were the bacterial strains we were planning to sequence, and we did. However, during the development of our project, two other studies reported genomes of M. pinnipedii3,4. They were used for the purposes of identifying ancient DNA from Peruvian mummies4, and to propose a novel taxonomic classification for the MTBC3. Therefore, with both MP1 and MP2 genomes in hands, we asked ourselves, why not go deeper into the understanding of these genomes? There were only few M. pinnipedii genomes available in public databases, but still, we could, at least, learn if MP1 and MP2 were identical and how they compared to other M. pinnipedii genomes deposited in databases.
Initiating a study exclusively on M. pinnipedii came as a surprise for us. As part of our Morris Animal Foundation Grant, our aim was to explore the genetic diversity of the Mycobacterium tuberculosis complex as a whole. To achieve this objective, we needed to sequence the genome of M. pinnipedii, because it had never been sequenced before. Our collaborators from the Laboratory of Bacterial Zoonosis at the College of Veterinary Medicine from the University of São Paulo had previously isolated two strains of M. pinnipeddi (MP1 and MP2) from a stranded sea lion carcass found in Capão da Canoa, Rio Grande do Sul (see map above)2. These were the bacterial strains we were planning to sequence, and we did. However, during the development of our project, two other studies reported genomes of M. pinnipedii3,4. They were used for the purposes of identifying ancient DNA from Peruvian mummies4, and to propose a novel taxonomic classification for the MTBC3. Therefore, with both MP1 and MP2 genomes in hands, we asked ourselves, why not go deeper into the understanding of these genomes? There were only few M. pinnipedii genomes available in public databases, but still, we could, at least, learn if MP1 and MP2 were identical and how they compared to other M. pinnipedii genomes deposited in databases.
Moving forward, the defined aims were to sequence and report the genomes of M. pinnipedii MP1 and MP2 and to perform a comparative genomic analysis of M. pinnipedii strains deposited in public databases. One of the cool parts of this study was the fact that we were able to include three ancient DNA sequences (1,000-year-old) of M. pinnipedii obtained from pre-Colombian Peruvian mummies provided by Bos and coworkers4 in some of our analyses. These ancient genome sequences were publicly deposited in GenBank.
What did the genomes of M. pinnipedii MP1 and MP2 tell us?
Our first analysis was aimed at identifying if M. pinnipedii MP1 and MP2 were identical, in other words, if they were the same strain infecting the sea lion. Species of the MTBC are clonal, very similar genetically, and have a slow mutation rate. They are not subjected to horizontal gene transfer and mutate solely through single nucleotide polymorphisms (SNPs), insertion sequence elements, deletions of up to ~15 Kb, and duplication of few gene families. MTBC species also often persist within individuals during a long-period of time, causing a chronic infection. These characteristics imply that a strain of MTBC can accumulate very few mutations over its co-existence with the host (usually in single digits – as few as 5-10), and this process is called microevolution. Alternatively, an individual can get infected with different MTBC strains through life (two or more infection events), and these strains will differ by a great number of SNPs. When this happens, an individual animal harbors a mixed-infection, also called “superinfection” (a fancy word to an infection occurring on top of an earlier infection, as two distinct transmission events).
Surprisingly, when we compared MP1 against MP2 we found a great number of SNPs and indels, which are characteristics of superinfection. We thus concluded that this sea lion was infected with two different strains of M. pinnipedii, constituting the first description of a mixed-infection in a sea lion. We also confirmed this finding using two DNA typing techniques traditionally applied for the MTBC: in silico spoligotyping and MIRU-VNTR PCR. We detected two spoligotypes and two loci of MIRU-VNTR with distinct patterns between MP1 and MP2. These were supportive evidence of a mixed-strain infection.
What is the importance of the detected superinfection in this sea lion?
Mixed-infections of M. tuberculosis in people normally occur in high-burden countries, where people are exposed multiple times to tuberculosis and where HIV plays an important role in shaping disease incidence. By tracing a parallel, we believe this sea lion was living within a population heavily infected with M. pinnipedii, allowing multiple chances of infection. Brazil does not have reproductive colonies of sea lions. Animals found at the coast are believed to have migrated from colonies in Uruguay. We decided then to search the literature for additional cases of tuberculosis in pinnipeds from South America. We found an interesting story happening in European zoos:
First, South American Sea Lions were imported from Uruguay to a zoo into Germany in 1992. Their offspring was sent to a zoo in Czech Republic, and later on these sea lions (captive-born) developed tuberculosis. Their parents (wild-caught) also developed tuberculosis in Germany, suggesting that the infection really came from the wild5. Outbreaks of tuberculosis in South American pinnipeds have also been reported in a French zoo6 (Video 1).
Video 1. Importation of infected South American Sea Lions into a European zoo and consequent disease spread to other zoos.
Another important study was conducted in Uruguay, in colonies of South American sea lions and fur seals (Arctocephalus australis)7. In this study, Arbiza and co-workers provide an elegant review of tuberculosis in pinnipeds worldwide, while reporting the disease as a concern for the survival of these species in Uruguay. Interestingly, the first isolation of M. pinnipedii in pinnipeds from Uruguay dates back from 1987, where 8/10 South American sea lions kept in a zoo were found with tuberculosis8. Since then, it seems that the detection of infected animals continues to occur, but its actual impact has not been extensively explored. Between 2001 and 2006, out of 154 evaluated pinnipeds (129 South American fur seals, 24 South American sea lions and one Southern elephant seal), five were found with granulomatous lesions in different organs, from which M. pinnipedii was isolated7. However, M. pinnipedii isolates were also obtained from organs without gross abnormalities from nine fur seals and the elephant seal (Mirounga leonine). Most importantly, as described by Arbiza and coworkers, Uruguay has prohibited exportation of South American sea lions in 2006 due to population decline9. It is plausible to believe this action has thus halted the continuous introduction of infected animals in zoos worldwide. However, according to the same authors, the exportation of wild-caught juvenile specimens of A. australis to zoos in Asia, Latin America and Europe still occurs. In the absence of strict control measures and effective diagnostic tests, such commercialization should be analyzed carefully.
Finally, just as we finished to write our manuscript, another Brazilian research group found an additional South American sea lion stranded on the coast of the same Brazilian State (Rio Grande do Sul)10. This animal was also sick due to M. pinnipedii infection and, unfortunately, did succumb to the disease while at care in the rehabilitation center. Therefore, we believe that, all these reports and the finding of superinfection in the sea lion of our study suggests that the problem of tuberculosis in South American pinnipeds may be larger than suspected.
What did we find when we compared all available M. pinnipedii genomes?
Unfortunately, not many genomes of M. pinnipedii have been sequenced in the world. In addition, data availability is not uniform; i.e. some genomes are available as paired-end reads, while others are available as single reads or draft genomes only. Therefore, not all comparative genomic analyses were performed with all available strains. We started by assembling M. pinnipedii genomes available as paired-end reads and annotating resulting contigs with RAST. We used OrthoMCL to find cluster of homologous proteins and we found that M. pinnipedii strains have a highly conserved proteome, with very few proteins reported as strain-specific or clusters within an accessory genome. This high conservation is expected in species of the MTBC, which are clonal bacteria, added to the fact that only few genomes were analyzed. We highlighted, however, that some hypothetical proteins and others associated with PE/PPE genes may be variably present in these strains. The problem is that, although RAST is a good annotation tool, it still reports truncated genes at the end of contigs (from draft genomes) as true genes. It also does not report on pseudogenes at all. An ongoing study in our laboratory show a fairly expressive number of pseudogenes in MTBC, which may thus skew analyses. Except for PGAP (from NCBI; not freely available), most commonly used annotation platforms also report these false genes at the end of contigs and do not report pseudogenes. Such discrepancies in gene annotation preclude correct interpretation of protein clustering analyses. Therefore, we recommended caution and future standardization of annotation platforms, as well as closing all sequence gaps, to provide better conclusions about genome variability of M. pinnipedii strains.
One important finding of our study was provided by our whole-genome based phylogenetic analyses. We showed that M. pinnipedii strains tend to cluster according to geographic location, and consequently, host species (as these animal species are already geographically segregated) (Figure 1). Such finding is just the tip of the iceberg to understand M. pinnipedii genomic diversity and transmission among animals of the same species and perhaps between species. As expected, the M. pinnipedii strains from the Peruvian mummies appeared emerging from a most basal node, suggesting ancestrality. Using these ancient sequences, we also detected a process of continuous genome reduction spanning up to 1,000 years, as MiD3 and MiD4 deletions could only be detected in modern M. pinnipedii strains. This finding supports previous observations of genome reduction in the MTBC as a whole. We also detected candidate deleted regions that still need to be confirmed using PCR or long-read sequencing in the future. Detecting deleted regions in MTBC genomes is always a challenge given the repetitive nature of their genomes and the size of the available reads.
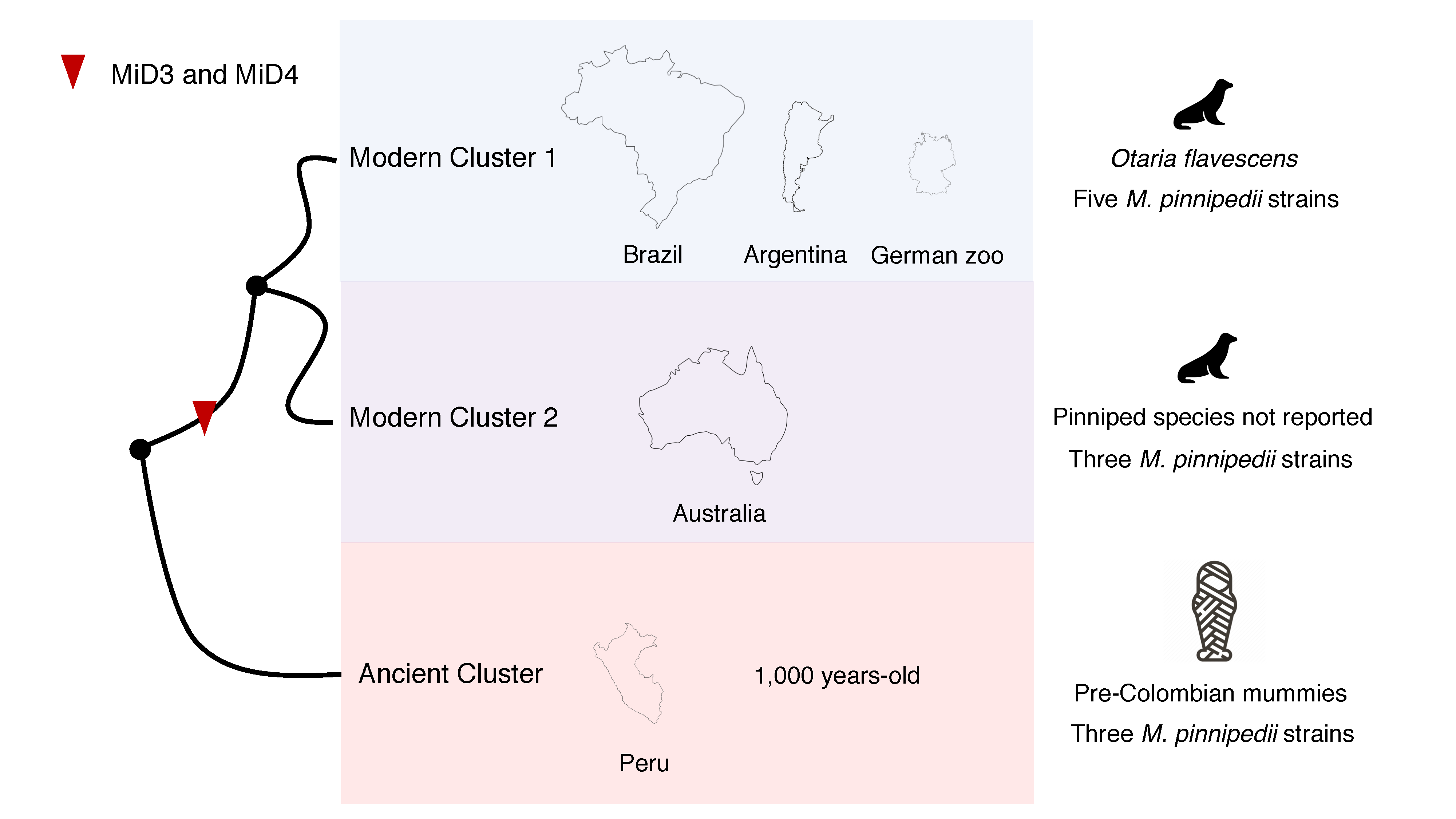 Figure 1. Schematic representation of Mycobacterium pinnipedii phylogenetic clusters detected with eleven strains of the bacterial species. This is not a phylogenetic tree. The phylogenetic tree can be found at Silva-Pereira et al., 2019.
Figure 1. Schematic representation of Mycobacterium pinnipedii phylogenetic clusters detected with eleven strains of the bacterial species. This is not a phylogenetic tree. The phylogenetic tree can be found at Silva-Pereira et al., 2019.
Take home messages and future perspectives
1) South American sea lions can be infected with more than one strain of M. pinnipedii.
2) Mycobacterium pinnipedii is likely to be highly endemic in the Otaria flavescens population from Uruguay.
3) This endemicity and the recurrent death of animals in the South American shores highlight the need of further investigations into the impact of tuberculosis in these animals.
4) There are at least two phylogenetic groups, cluster 1 and cluster 2, of M. pinnipedii circulating in South America and Oceania, respectively. Additional M. pinnipedii genomes must be sequenced globally as to understand their genetic diversity and association of distinct lineages/clusters with geography and animal species.
4) Genome annotation platforms need to be standardized to analyze MTBC genomes as to consider pseudogenes and/or gene prediction /annotation artefacts. These inconsistencies hamper comparative analyses into gene content and should be further addressed.
5) Mycobacterium pinnipedii is the only member of the MTBC to infect a marine mammal. Further comparative studies should be conducted to investigate variations in virulence factors and metabolism adaptation (mutations and gene loss or duplication) that allow this species to colonize an ocean animal and possibly survive in adverse environmental conditions permissive to its transmission.
References
- Silva-Pereira, T. T. et al. Genome sequencing of Mycobacterium pinnipedii strains : genetic characterization and evidence of superinfection in a South American sea lion (Otaria flavescens). 1–13 (2019).
- de Amorim, D. B. et al. Mycobacterium pinnipedii in a Stranded South American Sea Lion (Otaria byronia) in Brazil. J. Wildl. Dis. 50, 419–422 (2014).
- Riojas, M. A., McGough, K. J., Rider-Riojas, C. J., Rastogi, N. & Hazbón, M. H. Phylogenomic analysis of the species of the Mycobacterium tuberculosis complex demonstrates that Mycobacterium africanum, Mycobacterium bovis, Mycobacterium caprae, Mycobacterium microti and Mycobacterium pinnipedii are later heterotypic syn. Int. J. Syst. Evol. Microbiol. 68, 324–332 (2017).
- Bos, K. I. et al. Pre-Columbian mycobacterial genomes reveal seals as a source of New World human tuberculosis. Nature 514, 494–7 (2014).
- Kriz, P. et al. Case Report Mycobacterium pinnipedii in a captive Southern sea lion (Otaria flavescens): a case report Private veterinary practitioner, Czech Republic. Veterinarni Medicina 56, (2011).
- Jurczynski, K. et al. Pinniped Tuberculosis in Malayan Tapirs (Tapirus indicus) and its Transmission to Other Terrestrial Mammals. J. Zoo Wildl. Med. 42, 222–227 (2011).
- Arbiza, J. et al. Uruguayan Pinnipeds (Arctocephalus australis and Otaria flavescens): Evidence of influenza virus and Mycobacterium pinnipedii infections. New Approaches to Study Mar. Mammals. Tech Open, Rijeka, Croat. 248pp 151–182 (2012).
- Castro Ramos, M., Ayala, M., Errico, F. & Silvera, F. V. Aislamiento de Mycobacterium bovis en Pinnipedos Otaria byronia (Lobo marino comun) en Uruguay. Rev Med Vet 79, 197–200 (1998).
- Pèz, E. A. Situación de la administración del recurso lobos y leones marinos en Uruguay. (2006).
- Melo, A. M. et al. Tuberculosis caused by Mycobacterium pinnipedii in a wild South American sea lion Otaria flavescens stranded in southern Brazil. 133, 189–194 (2019).